Note
Click here to download the full example code
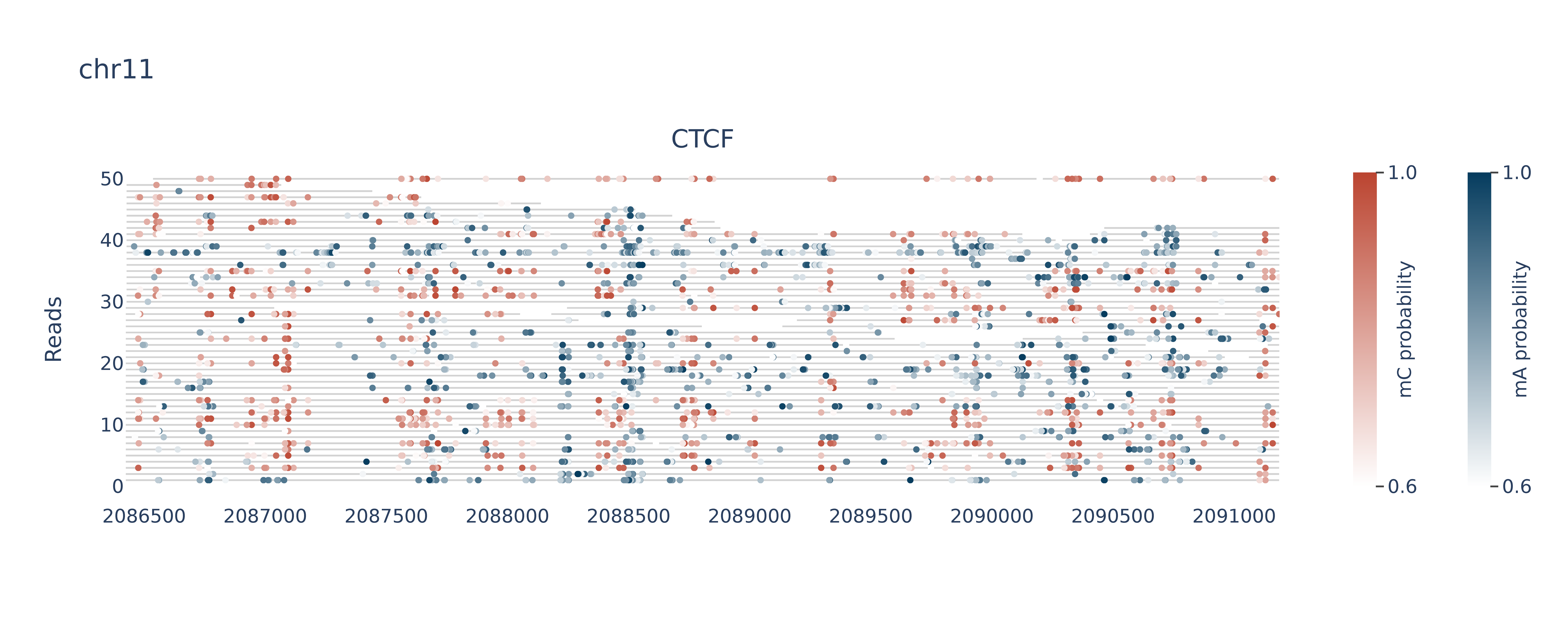
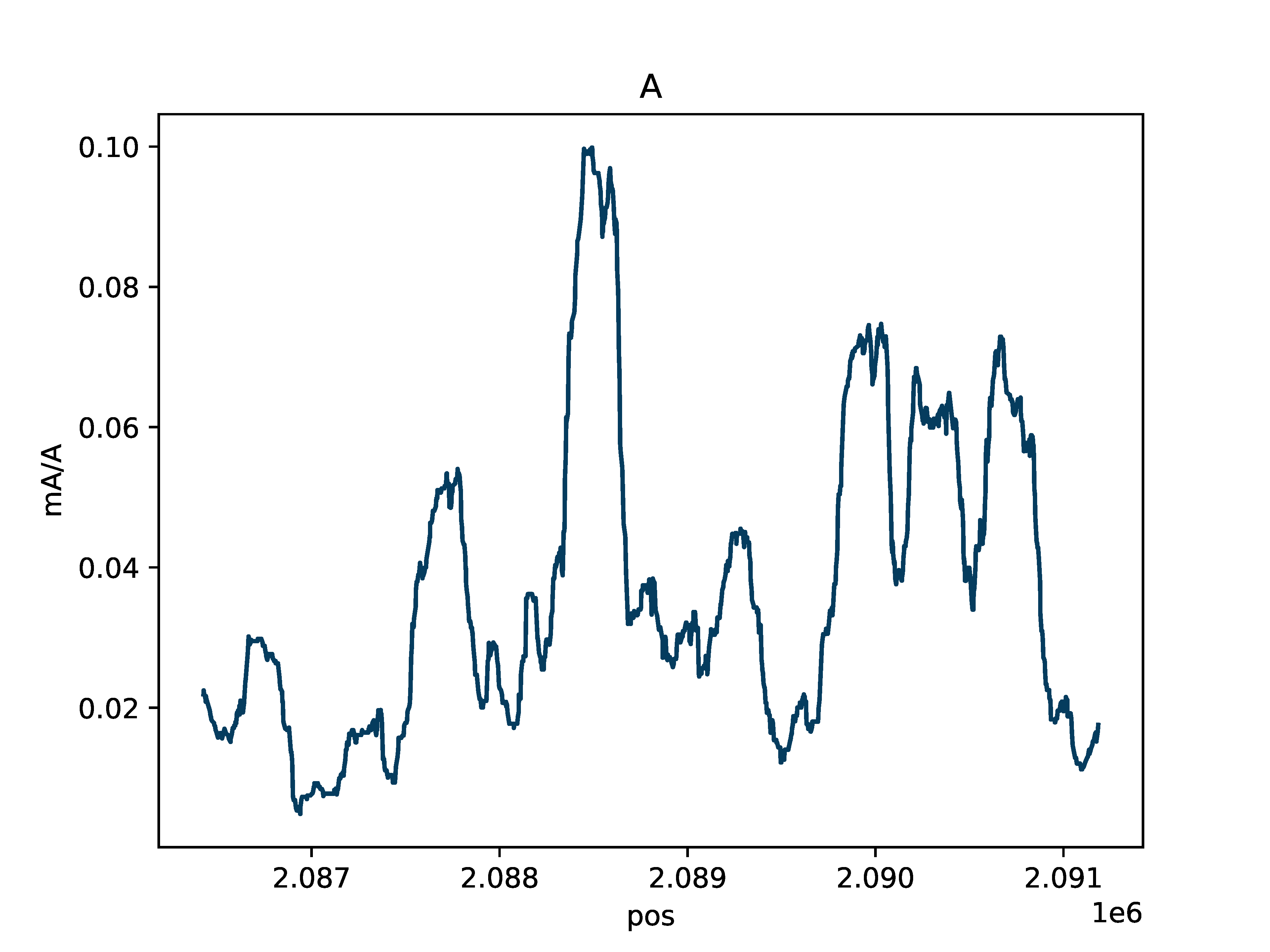
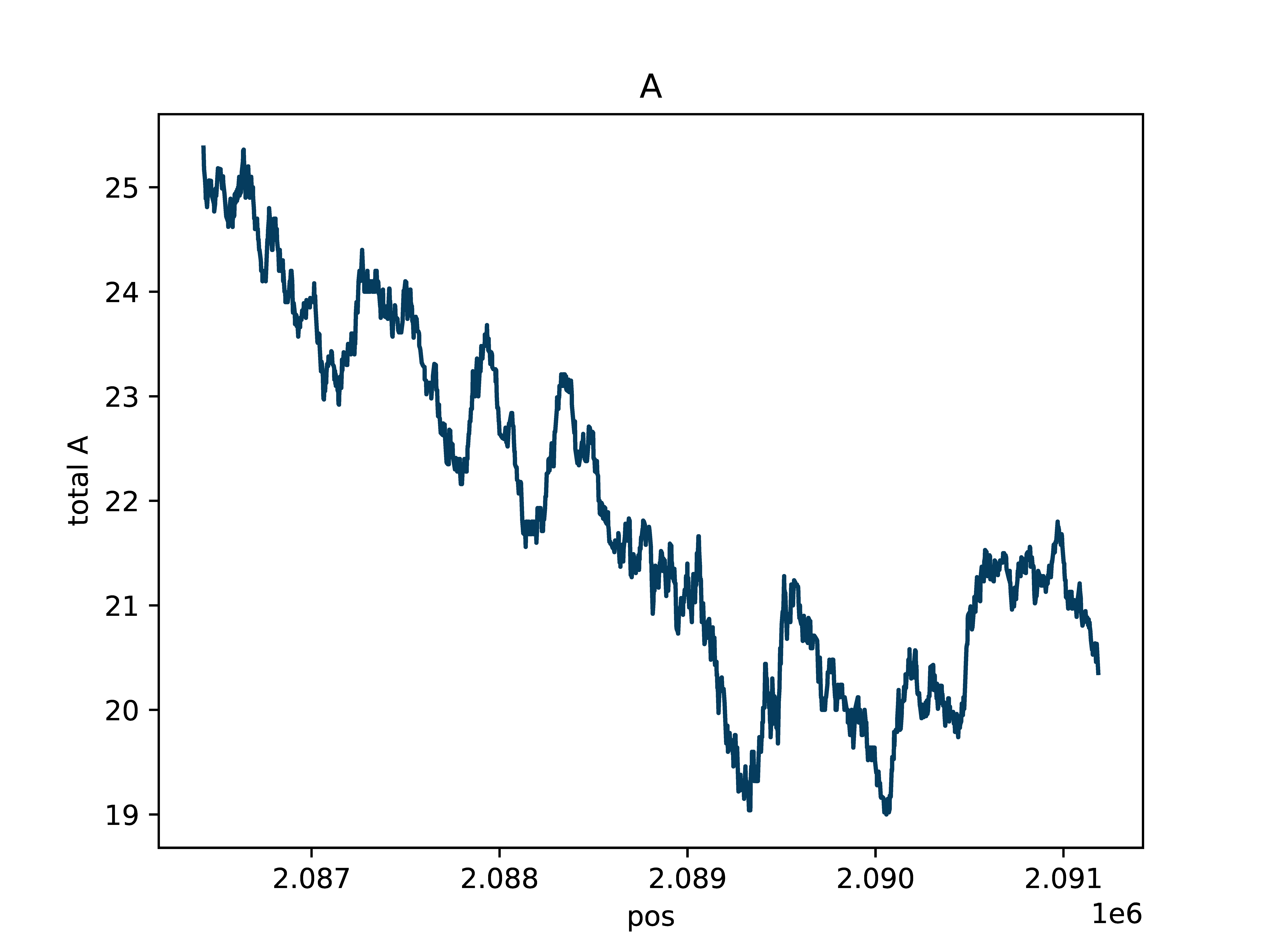
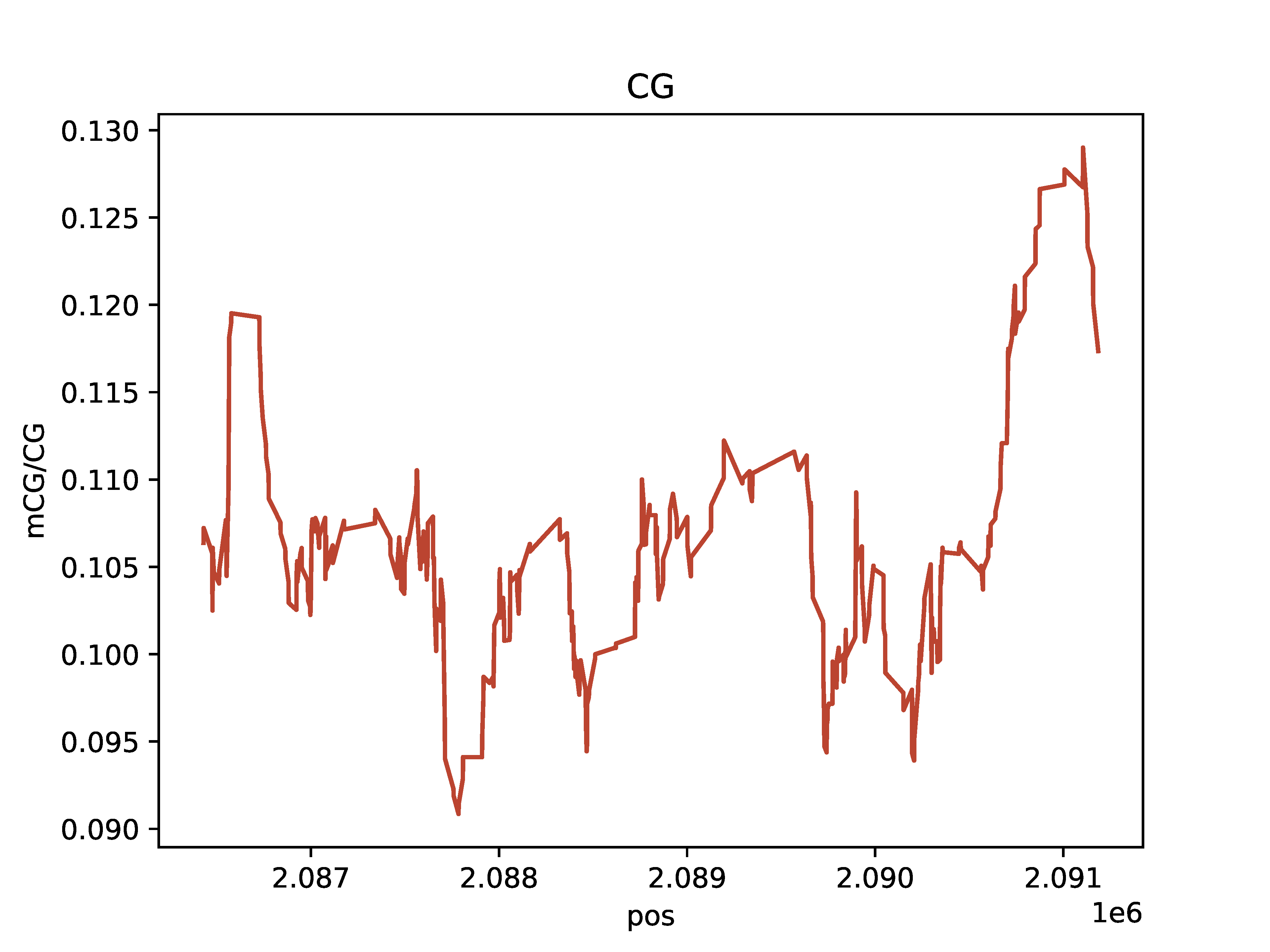
Single-Molecule Browser Plot
Plot single molecules with colored base modifications in a region of interest
Create either an interactive single-molecule HTML browser if static=False, or create a PDF if static=True. Base modifications are colored by type (mA vs. mCG) and probability of modification. Example data for producing these plots can be downloaded from SRA: https://www.ncbi.nlm.nih.gov/bioproject/752170 The below Python and command line options produce the same output.
1. Python option
import dimelo as dm
bam = "deep_ctcf_mod_mappings_merge.sorted.bam"
sampleName = "CTCF"
outDir = "./out"
dm.plot_browser(
bam,
sampleName,
"chr11:2086423-2091187",
"A+CG",
outDir,
threshA=153,
threshC=153,
static=True,
smooth=100,
min_periods=10,
)
2. Command line option
dimelo-plot-browser -f deep_ctcf_mod_mappings_merge.sorted.bam -s CTCF -r chr11:2086423-2091187 -m A+CG -o ./out -A 153 -C 153 --static -t 100 -n 10
Output
Total running time of the script: ( 0 minutes 0.000 seconds)