Note
Click here to download the full example code
QC Report
Outputs quality control report from given bam files.
Usually as a first step after receiving bam files, we want to do a quality check and assess our data. This code generates a PDF report of important QC statistics for modified base data from Nanopore sequencing.
1. Python option
import dimelo as dm
# first we specify the locations of our bam files
bam = "winnowmap_guppy_merge_subset.bam"
sampleName = "CTCF"
outdir = "./out"
# next we run the "qc_report" function
dm.qc_report(bam, sampleName, outdir)
# now our output directory will have a file called "CTCF_qc_report.pdf"
2. Command line option
dimelo-qc-report -f winnowmap_guppy_merge_subset.bam -s CTCF -o ./out
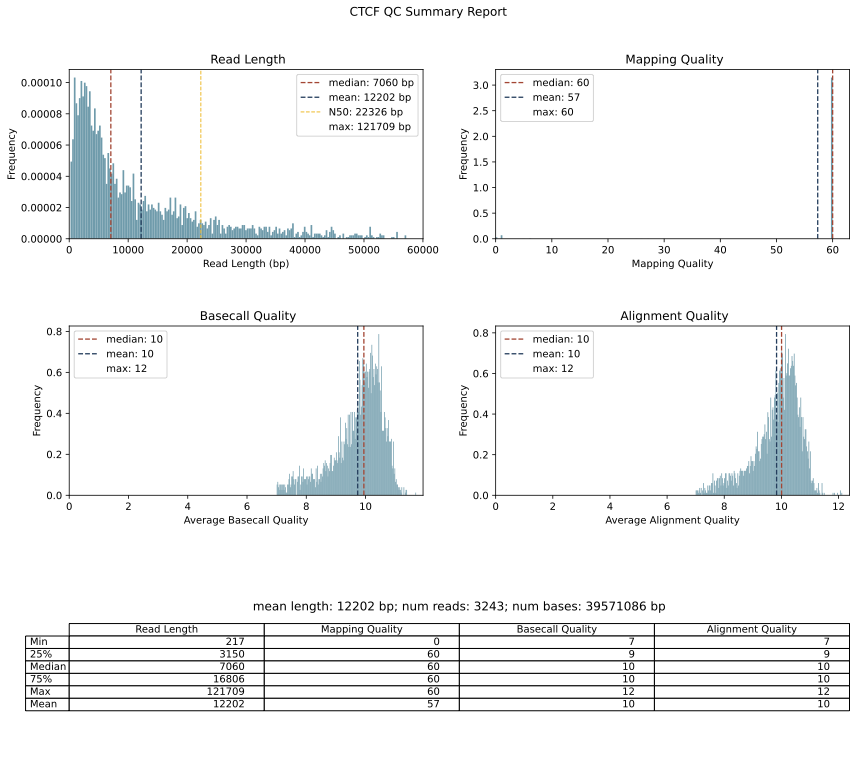
Output
Total running time of the script: ( 0 minutes 0.000 seconds)