Note
Click here to download the full example code
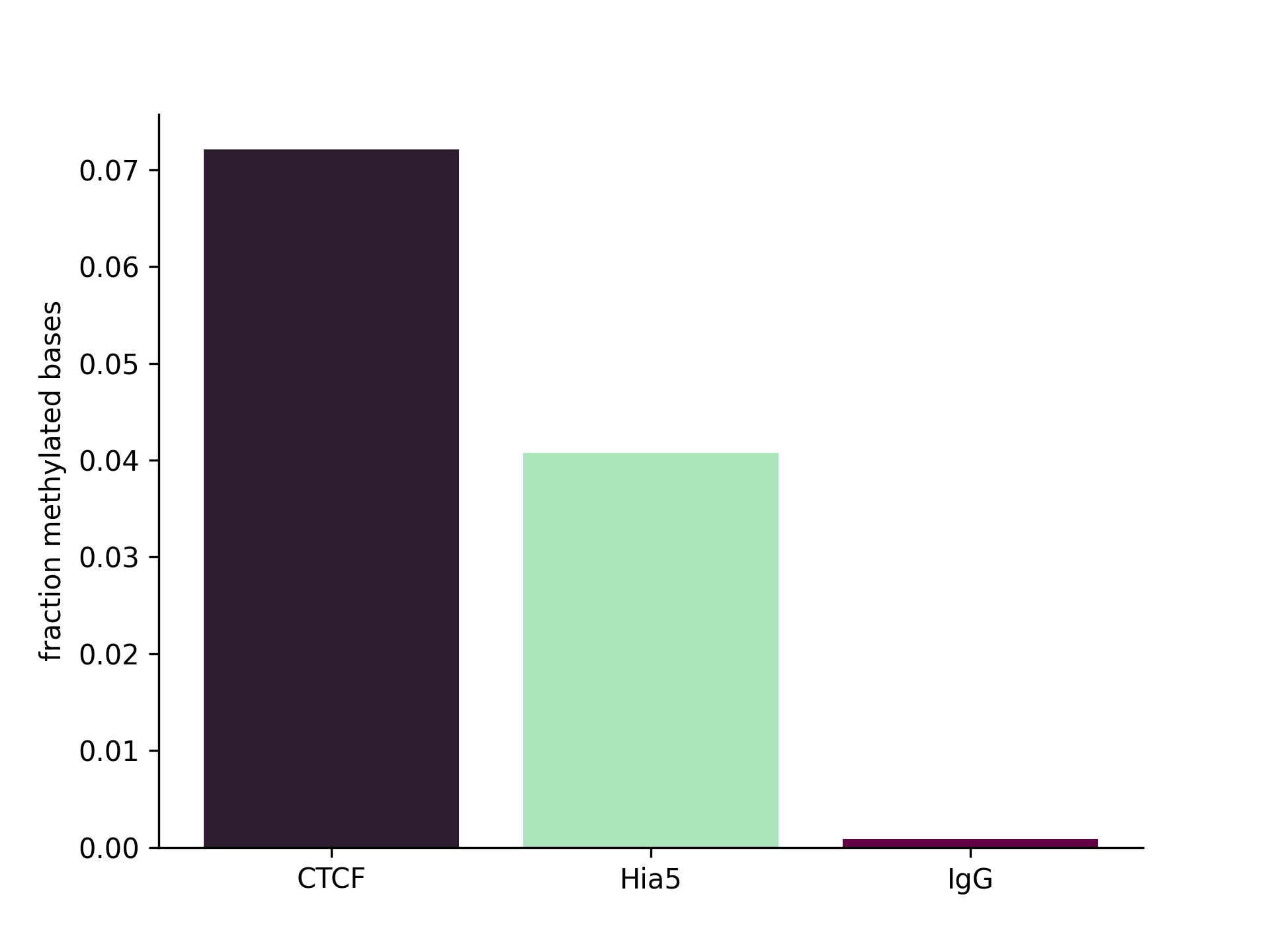
Enrichment Plot Comparison Across BAMs
Plot overall fraction of methylated bases within regions of interest specified by bed file across multiple samples.
Create barplot comparing methylation levels in bed file regions of interest across samples
1. Python option
import dimelo as dm
bams = [
"deep_ctcf_mod_mappings_merge.sorted.bam",
"hia5_mod_mappings.bam",
"igg_mod_mappings.bam",
]
sampleNames = ["CTCF", "Hia5", "IgG"]
bed = "q10.150.slop.bed"
outDir = "./out"
dm.plot_enrichment(bams, sampleNames, bed, "A", outDir, threshA=190)
2. Command line option
dimelo-plot-enrichment -f deep_ctcf_mod_mappings_merge.sorted.bam hia5_mod_mappings.bam igg_mod_mappings.bam -s CTCF Hia5 IgG -b q10.150.slop.bed -m A -o ./out -A 190
Output
Total running time of the script: ( 0 minutes 0.000 seconds)